
Botany online 1996-2004. No further update, only historical document of botanical science!
A direct relation exists between deoxyribonucleotide sequences (DNA) and the amino acid sequences of proteins. Nucleotide sequences carry the information that determines amino acid sequences, or, expressed the other way round, the amino acid sequence of a protein is encoded by a nucleotide sequence. The link between the two types of molecules that guarantees the transmission of the information is called the genetic code.
Deoxyribonucleotide sequences (DNA) consist of four different nucleotides (with the bases A, T, C and G), amino acid sequences of 20 different amino acids. The resulting question is: how does a code unit, a so-called codon, that encodes an amino acid look like and how many nucleotides does it contain?
One nucleotide is obviously not enough, since only four amino acids could be encoded this way. A pair of nucleotides (AA, AT, AG...etc.) would be insufficient, too, since they would result in 42 = 16 possible codons, not enough for 20 amino acids. So what about triplets? 43 = 64 codon possibilities exist here. That is not only enough but already too much.
Genetic experiments and physicochemical measurements suggested a triplet code. All codons are of equal length. But how are they organized? Are all 64 codons necessary? Do the codons overlap or not? Three theoretical alternatives exist:

The answer can be found by a simple consideration. In an overlapping code would an amino acid in a protein have an influence on the choice of the following amino acid. An amino acid that would be encoded by AAA, for example, could in a strongly overlapping code only be followed by an amino acid encoded by AAX, i.e. the first amino acid had always to be followed by one of just four different amino acids. In a weakly overlapping code (AAA would be followed by AXY) were only 16 (of the altogether 20) amino acids possible in the second position. The full potential of the 20 amino acids could thus only be used in a non-overlapping code.
Already in 1957 were enough experimentally determined amino acid sequences available to evaluate neighbourhood frequencies. S. BRENNER (Cambridge/ England) analyzed them and came to the conclusion that an overlapping code is out of question, since no amino acid in proteins was determined by its precursor.
Our language is an analogous example. In words can with one exception (a q is always followed by a u) each letter be followed by each of the other ones.
A further problem would be: how is the starting sign for the reading of a codon in a line of nucleotides given? - Interprete the figure above in a different way: Are the codons corresponding to the red, the blue or the green bars?
And finally now: which attempts existed to solve the genetic code?
A code can often only be solved, if the opponent makes mistakes and if the system behind these mistakes is understood. The genetic code is not free of mistakes. We know them as mutations. In the simplest case do they cause the exchange of an amino acid in an amino acid sequence. If the concept of the genetic code would be right then would one nucleotide have to be been exchanged by another in the respective nucleotide sequence.
Mutagenes causing specific and directed base substitutions in nucleic acids are known. Among them are nitrite ions (nitrous acid) that induce a transformation of C to U or of A to G by deamination.
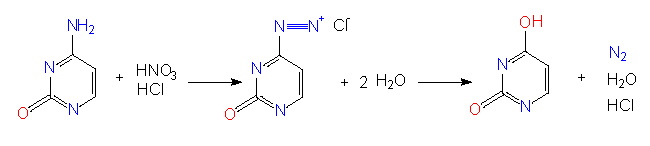
For biological experiments is the amount of such transformations important. The influence of nitrous acid on DNA (or RNA) should not last too long and its concentration should not be too high so that only a small percentage of the C and A residues in the nucleic acid are changed. This is a statistical process. We do know that Cs and As are effected but we do not know in which position. In many cases has such a modification an influence on information that is essential for life. As a rule causes therefore every mutagene an enhanced lethality and a large number of mutants can be expected among the few survivors.
Are such base substitutions induced by a mutagene mirrored in proteins as amino acid exchanges? A suitable test object was needed to answer this question and was found in the tobacco mosaic virus (TMV). The amino acid sequence of its coat protein is known since 1959. It consists of a sequence of 158 amino acids (analysis: G. SCHRAMM and his collaborators in Tübingen, A. TSUGITA and H. FRAENKEL-CONRAT in Berkeley). H. G. WITTMANN in Tübingen and A. TSUGITA and H. FRAENKEL-CONRAT in Berkeley produced a large number of nitrite-induced mutants, isolated single ones and analyzed the amino acid sequences of their coat proteins. It showed that certain amino acid sequences were changed compared to the wild type. The results can be summarized as follows:
The findings constitute a further proof that the code is not overlapping since two (three) neighbouring amino acids should have been changed in that case. Such cases were never found.
The exchanges were directed: the new amino acids were generally encoded by triplets richer in Us or Gs than the original codons.
The different exchanges could be arranged in a certain way that showed that several codons had to exist for a single amino acid. We would thus know what happens with the 64 - 20 = 44 'superfluous' codons. They are necessary, too. It is spoken of a degenerate code meaning that several codons exist for some amino acids (the term degeneration is used in the sense of redundancy).

Approaches to Decode the Genetic Code. The diagram shows exchanges yielded by the treatment of TMV with nitrite (diagram to the right). To the left are all those changes in the RNA shown that can be caused by treatment with nitrite, if UUU = Phe shall result in the end. See the text for further information (according to H. G. WITTMANN, 1962, 1966)
But how can it be decided in which position of a codon a C is chosen and where a U? To answer this a wholly different approach has to be chosen, one that led finally to the elucidation of the genetic code. It was found out how nucleic acids could be synthesized by free nucleotides. A. KORNBERG (Stanford University) isolated an enzyme (a DNA polymerase) that was able to produce the complementary strand with a single stranded DNA as a template. S. OCHOA (Rockefeller University, New York) isolated another enzyme (a RNA polymerase) that synthetizes RNA from ribonucleotides and needs no template at all. The offered triphosphate nucleotides are randomly polymerized to a polynucleotide chain. When offering only UTP or CTP are homogeneous sequences UUUUUU.. (= polyU) and CCCC... (polyC) respectively formed. When offering two nucleotides (like UTP and CTP) at the same time then are polymers produced that contain a random choice of Us and Cs.
What can such synthetic polynucleotides be used for? Not much, unless you have a system at your disposal that can read the contained information. M. NIERENBERG and H. MATTHEI (1961, National Institute of Health, Bethesda) developed an in vitro system able to perform protein synthesis. Necessary were: RNA, ribosomes, the supernatant of a bacterial extract (of Escherichia coli), amino acids as well as ATP, CTP, GTP and others. For the moment are two aspects important:
Protein synthesis was detectable under in vitro conditions. The test was quite simple: radioactively labelled amino acids were added and it was looked whether radioactivity could be precipitated after a short incubation with trichloracetic acid (TCA). It is known that free amino acids cannot be precipitated with TCA while proteins can.
After addition of a certain genetic information, like polyU could a polypeptide that contained only the amino acid phenylalanine (Phe) be precipitated. The first codon was solved: UUU= Phe.
If we return now to the mutants of the tobacco mosaic virus than we will see that Leu and Ser are encoded by codons richer in Cs (UUC, UCU, CUU) than that of Phe and that the C-content of the codons of Pro, Ser and Leu are be even higher (CCU, CUC, UCC or CCC). By comparison of further results gained with the in vitro system could also the exact sequence of the nucleotide bases in each codon be determined.
A conclusive answer to still open questions and the complete elucidation of the genetic code was possible when exactly defined (synthetic) polynucleotides of known base composition and -sequence were tested in the in vitro system mentioned above. In 1963 were the studies finished successfully.
The table shows the 64 codons and the amino acids they encode. The numbers 1, 2 and 3 refer to the position of the nucleotide within the codon, i.e. 1 = A, 2 = C, 3 = A: ACA = Thr. Three of the codons 'amber', 'ochre' and 'opal' signal a termination of the translation. A further codon, AUG, that codes normally for Met can also be the starting point for the peptide synthesis. The hydrophobic amino acids are placed in the upper left corner, the hydrophilic in the lower right one.
| 2 | |||||||||
| U | C | A | G | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | U | Phe | Ser | Tyr | Cys | U | 3 | ||
| Phe | Ser | Tyr | Cys | C | |||||
| Leu | Ser | ochre | opal | A | |||||
| Leu | Ser | amber | Trp | G | |||||
| C | Leu | Pro | His | Arg | U | ||||
| Leu | Pro | His | Arg | C | |||||
| Leu | Pro | Gln | Arg | A | |||||
| Leu | Pro | Gln | Arg | G | |||||
| A | Ile | Thr | Asn | Ser | U | ||||
| Ile | Thr | Asn | Ser | C | |||||
| Ile | Thr | Lys | Arg | A | |||||
| Met* | Thr | Lys | Arg | G | |||||
| G | Val | Ala | Asp | Gly | U | ||||
| Val | Ala | Asp | Gly | C | |||||
| Val | Ala | Glu | Gly | A | |||||
| Val | Ala | Glu | Gly | G | |||||
All 64 codons are used. 61 of them can be assigned to certain amino acids, the other three are stop signals. One of the codons can act both as an amino acid codon and as a start signal.
The different amino acids have different numbers of accompanying codons. For some, like Met or Trp exists just one codon, for others two or four and for some (Ser, Arg) even six. The frequency of the codons and the frequency of their amino acid is correlated. An exception is Arg, that has six codons but is underrated regarding its frequency in proteins.
The codons are not assigned randomly. The first two nucleotides of a codon have a higher informational value than the third one, GUU, GUC, GUA and GUG, for example, do all encode Val. Codons rich in UC encode hydrophobic, such rich in AG hydrophilic amino acids.
Many (nearly 30%) of all base substitutions do not change the encoding properties, for example:
UUU > UUC: Phe > Phe
Even if a base substitution causes an amino acid exchange is the chemical character of the side chain in most cases conserved (conservative exchanges). The genetic code can consequently be regarded as extremely conservative:
UUU > UUG: Phe > Leu
CUC > AUC: Leu > Ile
AAA > AGA: Lys+ > Arg+
AAA > GAA: Lys+ > Asp-
Exceptions exist (radical exchanges):
GAG > GUG: Glu- > Val
GAA> GUA: Glu- > Val
The last category leads usually to proteins that have lost their function or are impaired in it. Since they are subjected to selection have such mutants under natural conditions no or only reduced chances to survive.
During evolution has thus a genetic code evolved that guarantees stability and is fashioned in a way that a number of changes has no effect on the protein at all. The number of codons per amino acid, too, is not left to chance. It has already been said that frequent amino acids are encoded by several codons. But the questions what is cause and what effect remains. Without being able to answer this question exists at least a further correlation: amino acids with the costly syntheses have less codons than those where less energy is necessary for synthesis. The exception is again arginine.
Comparisons of the studies performed with the tobacco mosaic virus, micro-organisms and eucaryotic cells showed that the genetic code is universal, i. e. the assignments given in the table are the same for all living things. One exception was finally found: in animal mitochondria is the genetic information sometimes used in a deviant way due to a differing reading mechanism.
AUU: Met instead of Ile
AUA: Met instead of Ile
UGA: Trp instead of stop
AGA: stop instead of Arg
AGG: stop instead of Arg
In plant mitochondria, too, were variations found. Even species-specific differences seem to exist. In Oenothera causes UGA (TGA in DNA) a chain termination while CGG encodes Trp (W. SCHUSTER, A. BRENNICKE, 1985).
For more details see:
Now, how is the genetic information translated into a protein? Usually in a two-step process:
The information stored in the DNA is transcribed into RNA (transcription). Only those parts (genes) of the genome that are needed at a certain state are transcribed but never the whole genome at once.
The (partial) information now contained in the mRNA is translated into a protein in a complex process involving a multitude of components (translation or protein biosynthesis).
© Peter v. Sengbusch - Impressum