As their name suggests, amino acids are compounds that contain both an amino group and a carboxylic acid unit. The amino acids of interest to most people are a-amino acids, which means that the amino group is attached to the carbon atom that is a to the carboxyl carbon. A generalized structure for a-amino acids is shown in Figure 1 where R represents any of the more than 20 substituents that are found in naturally occurring amino acids.
There are several structural features that are characteristic of amino acids. First, they exist as doubly charged ions called zwitterions. One piece of evidence that suggests the ionic nature of amino acids is their high melting points. Alanine (R=CH3), for example, melts at 315oC. By comparison, 2-amino-1-propanol is a liquid that boils at approximately 175oC. Clearly the intermolecular forces between alanine molecules are much greater than the dipole-dipole interactions that hold molecules of 2-amino-1-propanol together.
With the exception of glycine, where R = H, Ca is chiral in all of the amino acids. Furthermore, all the chiral amino acids have the same absolute stereochemistry at Ca. They are all designated as L-amino acids. This designation is based upon the stereochemistry in glyceraldehyde. Figure 2 uses Fischer projections to compare the amino acids with D-glyceraldehyde.
According to the IUPAC convention, the correct name for L-(+)-alanine is S-(+)-alanine. While all of the chiral amino acids have the same absolute stereochemistry, they are not all dextrorotatory. Phenylalanine (R = CH2C6H5), for example, is levorotatory, i.e. it is L-(-)-phenylalanine or S-(-)-phenylalanine. Just to confuse you, L-cysteine (R = CH2SH) is dextrorotatory and its IUPAC name is R-(+)-cysteine. (Why R and not S?) Interactive models of L-(+)-alanine and L-(+)-cysteine are shown in Figure 3.
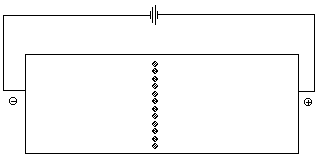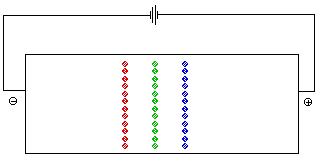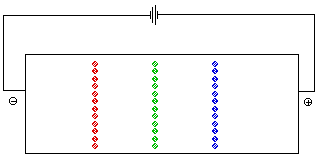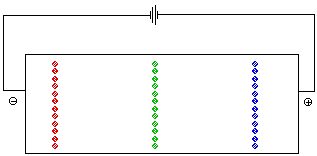
Most amino acids are obtained by the hydrolysis of proteins. The process produces mixtures. One technique for separating the individual amino acids is called ion exchange chromatography. Another method that is frequently employed to monitor the composition of a mixture of amino acids is called electrophoresis. In order to undestand how these approaches work it is necessary to consider how the net charge on an amino acid varies as a function of pH, i.e. its pH profile. Figure 4 illustrates the change in charge as the pH of a solution of lysine is increased from 1 to 12.
Lysine contains a carboxylic acid group, the a-amino group, and a second amino group attached to the side chain. In a very acidic solution, pH=1, all of these groups are protonated. The net charge on the lysine is +2 (State A). As the pH of the solution is raised, by the addition of NaOH for example, the most acidic site in lysine will be deprotonated first. This is the carboxylic acid group. The pKa of the CO2H proton is 2.2. This means that when the pH of the solution reaches 2.2, 50% of the CO2H groups in lysine will be deprotonated. More NaOH will deprotonate the remaining CO2H groups until 100% of the lysine is present in State B. The net charge in State B is +1. As the pH increases, the NaOH will begin to deprotonate the a-aminium group which has a pKa of 9.0; at a pH of 9.0, 50% of the a-aminium ions will be deprotonated. The net charge on a lysine molecule in State C is 0. Once all of the a-aminium groups have been deprotonated, the NaOH will deprotonate the aminium group in the side chain. The pKa of this group is 10.5. When the pH equals 10.5, 50% of these groups will be deprotonated. Further addition of NaOH will deprotonate the remaining aminium groups in the sample. The net charge on a lysine molecule in State D is -1.
The pH at which the net charge of an amino acid is 0 is called the isoelectric point. It is represented by the symbol pI. At a given pH the net charges on amino acids with different pI values are different. Both ion exchange chromatography and electrophoresis depend on differences in the net charge on a molecule in order to separate the components of a mixture. In an electrohoresis experiment a sample is placed at the center of a bed of an electrically conductive gel . If a volatge is applied across the gel, positively charged ions migrate toward the cathode (the negative electrode) while negatively charged ions move toward the anode. Molecules with no net charge do not move. Figure 5 animates the separation of a mixture of glycine, lysine, and aspartic acid at a pH of 6. At this pH the net charge on the aspartic acid is -1, while that on the lysine is +1. There is no net charge on the glycine.
Peptides are amides formed by the reaction of the a-amino group of one amino acid with the carboxylate group of another amino acid. Peptides which contain two amino acids are called dipeptides. Figure 6 shows the structure of a dipeptide formed from glycine and alanine.
Peptides made from three amino acids are called tripeptides, etc. Peptides containing from 2-10 amino acids are arbitrarily called oligopeptides. Compounds containing more amino acids are called polypeptides or proteins.
In order to simplify the representation of peptides, chemists assigned a 3-letter code to each amino acid. By this convention the dipeptide in Figure 6 would be Gly-Ala; the N-terminal amino acid is written first. Bradykinin, a nonapeptide which is involved in regulating blood pressure, has the structure Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe-Arg.
Peptides are structurally complex. Their structural features have been broken down into 4 categories.
This is simple. The primary structure of a peptide is the sequence in which the amino acids are connected, starting with the N-terminal amino acid.
The primary structure of a peptide doesn't convey any information about the 3-dimensional shape of the peptide. The secondary structure does. In talking about secondary structure, we are actually referring to the topology of regions within the peptide. Such localized structure is a reflection of the conformations about a sequence of peptide bonds. These conformations are determined in large part by the nature of the R groups attached to the a carbon. Biochemists have identified three common topologies, helices, pleated sheets, and turns. Figure 7 presents two interactive models of Gly-Gly-Gly-Gly-Gly-Gly-Gly-Gly-Gly which clearly indicate the helical nature of this synthetic nonapeptide.
The tertiary structure of a polypeptide describes the way in which the chain loops and twists and bends. While the primary structure usually depicts the backbone of a polypeptide as an extended chain in which the N-terminal amino acid is far away from the C-terminus, the fact is that the chain is not extended, and it is possible for the 5th amino acid to be spatially quite close to the 45th or the 450th, for example. Figure 8 shows the structure of one polypeptide chain from human insulin. This chain contains 51 amino acids.

One of the roles that proteins play is that of reaction catalyst. In this role proteins are more commonly referred to as enzymes. Most enzymes are comprised of two or more polypeptide chains that are held together by non-covalent forces. The individual polypeptide chains are called sub-units. Hemoglobin, for example, contains four sub-units, each of which is organized around a central iron atom. Human insulin is a hexamer, i.e. it contains 6 sub-units. The spatial relationship of one sub-unit to another is called the quaternary structure of a protein.
Proteins serve two main functions, structural and catalytic. Structural proteins are generally fibrous in nature. Hair and smooth muscle are examples of structural proteins. Catalytic proteins generally have a globular quaternary structure that is more or less spherical. The catalytic site is buried in the interior of the structure.