
Botany online 1996-2004. No further update, only historical document of botanical science!
Some bacteria and blue-green algae are able to reduce atmospheric nitrogen to ammonia. Several of them do even live in symbiosis or in association with green plants. The nodule bacteria (e.g. Rhizobium) of legumiosae are best known. They are host-specific. Rhizobium japonicum lives in symbiosis with soy beans, Rhizobium trifolii with clover, Rhizobium meliloti with lucerne. Anabaena azollae (a blue-green alga) co-operates with an aquatic fern, Nostoc muscorum (another blue-green alga) with the tropic plant Gunnera macrophylla. The leguminous genus Pisum contains species living in continuous symbiosis with nodule bacteria, others that develop non-functional nodules, and finally such species that do not grow nodules at all, being consequently unable to form symbioses.
A number of free-living soil bacteria, e.g. bacteria of the genera Azobacter (aerobic), Closterium (strictly anaerobic), Klebsiella (optionally aerobic), and Rhodospirillum (anaerobic, photosynthetically active) belong to the nitrogen reducing species.
Nitrogen fixation has been thoroughly covered during the last years, since genetic engineering fosters the hope for techniques improving the nitrogen supply of plants. The production of synthetic nitrogen fertiliser is expensive and extraordinarily costly in terms of energy. Bacteria, too, are not able to produce ammonia at low energy costs, since the triple bond of nitrogen belongs to the strongest covalent bonds occurring in biologically important molecules. The conversion of 1 mole nitrogen to 2 mole ammonia requires 25 mole ATP, i.e. the fixation of 1 gram nitrogen costs 10 g glucose - under favourable conditions. Azotobacter’s reaction is especially pricey: it needs 100 g glucose for the fixation of 1 g nitrogen.
The genetic basis of nitrogen fixation is largely known. The preferred test object was and still is Klebsiella pneumoniae, an enterobacterium belonging to the kinship of Eschericia coli and the salmonellas. In nitrogen fixation, the nitrogenase complex takes up a key position. The encoding and the regulation of this protein is controlled by a certain DNA region, the nif-region, that contains 16 (or 17) genes in the case of Klebsiella. The nif-genes belong to seven different operons (transcription units). Except for one gene that is located on the complementary strand, all of them are located on the same (the ‘encoding’) strand.
Mo nitrogenase component I - reduced and oxidized (upper row) - Biological nitrogen fixation, i.e. reduction of molecular nitrogen to ammonia (1), is catalysed by the nitrogenase enzyme system (EC 1.18.6.1). Molybdenum nitrogenase (Monitrogenase), which is found in all nitrogen fixing organisms, consists of two components: component I [nitrogenase molybdenum-iron (MoFe) protein, or dinitrogenase], and component II [nitrogenase iron (Fe) protein, or dinitrogenase reductase] [1; see list of reviews on structure and function of Monitrogenase]. © PROMISE
In Azobacter, the genes are scattered over the whole genome. The nif-region of Klebsiella has been isolated, cloned, and expressed in Eschericia coli. Nevertheless, the transformation of green plants poses several principal problems:
The genes have to be coupled to a eucaryotic promoter in order to be expressed.
Oxygen-free compartments or zones are required, since nitrogenase is extremely sensitive towards oxygen.
The electron transport chain of the plant cell has to be in tune with that of the nitrogenase, and finally.
have sufficient amounts of ATP to be provided.
The problem is thus not the transformation of the plant cells or the integration of the foreign genes into the plant genome, but to obtain control of both their expression and the activity of their gene products. No practicable solutions exist as yet. It is rather tried to increase the efficiency of soil bacteria or to optimize the conditions of the plants’ rhizosphere.
So much for the bacterial side. And how do the plant species apt for symbiosis with nitrogen-fixing bacteria contribute to the symbiosis? How are – to be more detailed – the following problems covered?
- How do bacteria and plant root recognize each other in order to establish the following interaction ?
- How does the bacterium infect the plant cell ?
- Which plant genes are activated after infection ?
- Which changes occur in the bacterium ?
- How is the ammonia produced by the bacteria used ?
It is well-known that most cellular surfaces are studded with carbohydrates. Consequently, the carbohydrate patterns of bacterial surfaces and of root hair surfaces were screened for a possible participation in the interaction of bacterium and plant. Moreover, plants do produce carbohydrate-binding proteins, the lectins, that could act as linking elements. This model assumption was verified by specific experiments. F. B. DAZZO and D. H. HUBBELL (Michigan State University) proved in 1975 that Trifolium repens (white clover) secretes a lectin (trifolin) with an affinity for 2-desoxyglucose. This sugar occurs both at the cell surfaces of Rhizobium trifolii and at the root hairs of Trifolium-species. Rhizobium trifolii-mutants lacking 2-desoxyglucose exist. They are unable to infect clover or to induce the production of nodules.
Rhizobium japonicum is characterized by its exposed galactosyl-residues. The lectin of its host plant, soy bean agglutinin (SBA) reacts specifically with galactose.
Nodule bacteria are mobile. They find the rhizosphere of plants chemotactically. Phenolic compounds secreted by the roots, e.g. luteolin, serve as signals. They activate the NodD gene of the nodule bacterium that regulates further genes of the ‘nod-box’: nodA, nodB, and nodC. These genes occur in all nodule bacteria (common nod-genes), and a further group of nod-genes (nodH...) participates in the reaction between bacterium and host plant. The production of an additional signal molecule is followed by the first visible morphological change: the root hairs start to bend.
Bacteria of the genus Bradyrhizobium have a two-component system: nodV and nodW. Here, too, both components seem to be necessary for nodulation. Besides the nod-genes, a further group of genes, the hsn genes (host specificity of nodulation) are known. They display their effects only in their original species, i.e. they are unable to complement the respective genetic defect of another bacterial species.
LEROUGE et al. discovered in 1990 that Rhizobium meliloti, a nodule bacterium living in symbiosis with alfalfa (lucerne), produces a further, highly specific molecule, nodRm-1. NodRm-1 seems to elicit root hair deformation directly. The structure of nodRm-1 is new. It is a (beta)-linked glucosamine-tetrasaccharid. Three of the four sugar groups are N-acetylated. A sulphate group sits at the reduced end, while the non-reduced end carries an N-acetyl-C16 fatty acid. The molecule resembles no known intracellular phytohormone or regulator molecule. This is of special interest, since oligiosaccharides have been paid quite a lot of attention recently, when it was discovered that certain members of this molecular group participate in the specific interaction of plants and fungi and act as regulators of differentiation processes.
Infection of a plant with Rhizobium causes the development of nodules. The synthesis of two plant proteins, nodulin and leghaemoglobin, is especially interesting. Nodulin causes enlargement and multiplication of the cortical cells. During these changes, the degree of ploidy of the nuclei rises, too. The single steps of the causal chain are less well known. It is not known, whether the enlargement of the cells is caused by polyploidy or whether it is a direct effect of nodulin. The nodular tissue has a blood-red colour. It contains leghaemoglobin, a protein homologous to animal haemoglobin. Their amino acid sequences are similar, the tertiary structures largely identical. This led to the assumption that globin genes existed even before animals and plants developed. Accordingly, they would have to be very old genes and the question, why they are only sporadically expressed in the plant kingdom remains unanswered.
The DNA analysis showed that leghaemoglobin contains four exons, haemoglobin only three (M. GO, 1981), indicating that the single fragments are usually scattered over the genome. Haemoglobin, respectively leghaemoglobin synthesis occurs only, if the exons are united as one transcription unit by translocation. In the plant kingdom, this occurred in leguminosae and in Parasponia rigida, a species from the Malayan archipelago that is related to elms (C. A. APPLEBY et al., 1983). Just like in the case of animal haemoglobin, only three exons exist in these cases. Meanwhile, haemoglobins have been detected in roughly a dozen different, non-related plant families. The function they have in these plant species remains so far unknown.
The protein component of leghaemoglobin is encoded by the plant genome, while the production of the porphyrin ring occurs within the bacterioid. Accordingly, bacterial genes participate in the synthesis of the porphyrin ring whose activity has to be in tune with that of the plant globin genes. The central iron ion of the porphyrin ring is again provided by the plant. The bacterioids need quite a lot of iron ions as well as molybdenum ions since both are essential for the activity of the nitrogenase. Nitrogenase is sensitive towards oxygen. Leghaemoglobin binds to oxygen and generates thus oxygen-free areas within the roots of plants where the bacterial nitrogenase can become active.
Nodule bacteria, too, change during their synthesis with plants. They become bacteroids that lack the outer membrane. The cells branch. The degree of branching decreases with age.
Nitrogenase produces ammonia, a heavy cytotoxin, that the bacteria pass on to the plant cell.

The ammonia can be processed in at least three ways that render it harmless and at the same time make it usable in a bound form as amino acids.
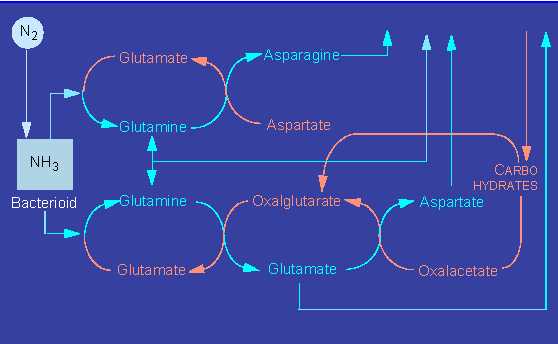
The plant enzymes participating in these processes are glutamate dehydrogenase, glutamine synthethase, and glutamate synthethase. In addition, the plant has to supply sufficient amounts of carbon skeletons as acceptors in the case of all three pathways.
Although extremely important, the contribution of symbiotic bacteria to the nitrogen supply of plants is very small when seen in relation to nature’s total nitrogen budget. The amount of cultivated leguminosae is less than 10 percent of all cultivated plants: cereals are in the front position. In some species, e.g. sugar cane and rice, an accumulation of nitrogen-binding bacteria has been detected in the rhizosphere. Plant roots secrete up to 20 percent of the carbohydrates produced by photosynthesis. The bacteria are able to use them as acceptor molecules for ammonia.
In some cases, that of Spartina alternifolia or several rice species, for example, the bacteria penetrate the intercellular spaces of the root tissue. The co-operation between plant and bacteria works especially well with C4-plants, though some C3-plants do host bacteria, too. Azospirillum brasiliense, for example, is a bacterium that associates almost exclusively with C3-plants. It is dependent on the supply with organic acids, namely malate. Data about the amount of nitrogen gained by the cultivation of grasses (Gramineae) exist. They show that the highest gains are made in the tropics.
The interactions of Anabenae-Azolla symbioses differ from that of leguminosae-nodule bacteria interactions. Little is known about the way in which Anabena and Azolla recognize each other. Anabaena enters the fern’s tissue at tip of growing shoots. Nitrogen fixation takes place in specialized cells, the heterocysts, that alternate with vegetative, photosynthetically active cells in the alga’s filaments. Roughly every tenth cell is a heterocyst. In the case of the Anabaena-Azolla interactions, the penetrating Anabaena cells are small, and heterocysts are lacking. Heterocyst development starts not before Anabaena has colonized the fern tissue and has settled within the intracellular cisterns (H. D. HILL, 1977). Azolla is common in the rice fields of Eastern Asia, where a considerable amount of the nitrogen bound by this fern is of benefit to the rice plants.
Symbiotic and free living Anabaena-species have – just like other blue-green algae, too – to face the problem of protecting themselves against oxygen. On one hand, metabolic processes intercepting surplus oxygen exist, on the other hand, heterocysts surrounded by bacteria have often been observed. In contrast to vegetative cells, active heterocysts are enclosed by a coat of polysaccharides that seems to serve as a nutrient for bacteria. The metabolic activities of the bacteria again consume oxygen, thus generating low-level oxygen microzones around the heterocysts.