Botany online 1996-2004. No further update, only historical document of botanical science!
Chloroplasts are the typical organelles of green plants. They are the site of photosynthesis though they perform a number of further synthetic processes, too.
Chloroplasts (and other plastids, like leucoplasts, amyloplasts or chromoplasts) develop either by division of an existing plastid or from proplastids. Proplastids are a stage of differentiation that arises during germ cell formation. When exposed to light do they develop into ripe plastids.
Without being exposed to light the inner membranes form a lattice-like prolammellar body that transforms into the thylacoid system after illumination. In unicellular algae (Euglena, Chlamydomonas) and in higher plants, e. g. barley (Hordeum vulgare), a number of mutants has been described with different blocks of the chloroplast synthesis. By studying such mutants it was possible to analyze the individual steps in chloroplast synthesis and to determine their sequence. In green tissues there is a close correlation between the division rate of the chloroplasts and the mitotic rate.
In contrast to the compartments and membranes talked about so far are the chloroplasts of many plant species easily isolated. Isolation is usually achieved by density gradient centrifugation or, not that popular any more, by differential centrifugation. This is the reason why we know pretty much about chloroplast membranes and processes.
Chloroplasts have three principally different types of membranes:
![]() The outer membrane. Its composition resembles that
of other cytoplasmatic membranes.
The outer membrane. Its composition resembles that
of other cytoplasmatic membranes.
![]() The inner membrane and finally
The inner membrane and finally
![]() the thylacoid membrane where photosynthesis takes place.
the thylacoid membrane where photosynthesis takes place.
Scheme characterizing the membrane surfaces after freeze etching. The expressions are based on a nomenclature suggested by BRANTON et al. P: surface facing the cytosol (stroma); E: surfaces facing the exoplasmatic lumen; F: fracture within the membrane; S: surface of the membrane; s: stacked membranes; u: unstacked membranes (according to L. A. STAEHELIN, 1976)
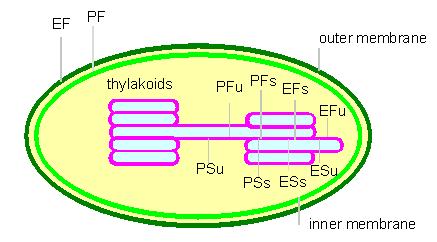
The outer and the inner membrane together are usually called the chloroplast envelope. The thylacoid membranes develop by invagination and subsequent pinching off of the inner membrane. They are grouped into grana (stacked) and stroma (unstacked) parts. Information about the involvement of the different membrane sections can be found under:
The cells of the bundle sheaths of C4 plants (sugar cane, maize, etc.) contain incomplete chloroplasts where only partial photosynthesis takes place (PS I). Stroma and grana can be reversibly transformed into each other. A missing activity in stroma thylacoids does not mean that the required enzymes are missing, too. It was shown that they exist, but that they are not combined in functional units (oligomerous complexes). The activities are controlled by the aggregating behaviour of the subunits and the co-operative interactions of membranes on top of each other.
Since 1973 have the chemical composition and the properties of the chloroplast membranes mainly been studied in the laboratory of R. DOUCE. They contain an enzyme system, that transfers galactosyl subunits from uridine diphosphate galactose onto diglycerides. The resulting monogalactosyl diglycerides and digalactosyl diglycerides are incorporated into the chloroplast membranes and are a typical feature. Moreover do the two outer membranes contain the xanthophyll violaxanthin that lends them a slightly yellowish colour. Chloroplast envelopes that have been exposed to strong light become more orange due to a conversion of violaxanthin to zeaxanthin.
Chloroplasts contain genetic information. The significance of their genes was first understood in the early works of E. BAUR (1909) and C. CORRENS (1909). The molecular genetic phase of research started in 1962 with the definitive proof of DNA in chloroplasts (H. RIS and W. PLAUT, University of Wisconsin, Madison). Chloroplast DNA (ctDNA) is circular. Although it contains information about the formation of a whole lot of chloroplast proteins exist by far more proteins in chloroplasts than encoded by the ctDNA:
100 genes alone would be required for the DNA and protein synthesis in chloroplasts (DNA polymerases, RNA-polymerases, rRNA, ribosomal proteins, tRNA, aminoacyl-tRNA-synthethases, soluble factors). 40 additional enzymes are needed for the synthesis of chlorophyll and carotenoids and again 40 enzymes take part in the other activities of photosynthesis. As far as the figures go could the chloroplast DNA contain just enough genes for all these enzymes. In nearly all examined higher plants does it contain about 150 000 base pairs. But the list of the chloroplast activities is not finished yet. Furthermore are
![]() enzymes for lipid and amino acid syntheses,
enzymes for lipid and amino acid syntheses,
![]() enzymes for the carbohydrate metabolism and
enzymes for the carbohydrate metabolism and
![]() Enzyme enzymes for the synthesis of a number of secondary plant products
.
Enzyme enzymes for the synthesis of a number of secondary plant products
.
| C N O Cl |
required. The list shows that quite some enzymes have to be imported from the cytosol into the chloroplasts. An equilibrium of chloroplast encoded and nuclear encoded functions exists. The enzymes required for the replication of the ctDNA, for example, are encoded by the nucleus. They are synthesized in the cytosol and are afterwards imported into the chloroplasts. The chloroplast ribosomes contain rRNA encoded by the chloroplasts and proteins of which some are encoded by the chloroplasts themselves while others are encoded by the nucleus. Whether a protein is synthesized in the cytosol or in the chloroplasts can be detected by the use of specific inhibitors (antibiotics). Streptomycin and chloramphenicol (see also picture to the left) inhibit the protein biosynthesis at the ribosomes of chloroplasts (and that of procaryotes). Cycloheximid has no influence on the protein synthesis of chloroplasts but it prevents that of the cytosol. The origin and the site of synthesis of a number of proteins could be determined using these inhibitors.
Further important for answering this question was the use of mutants (for example Euglena gracilis and Chlamydomonas reinhardii). A genetic analysis shows whether the defect has occurred in the chloroplast or the nuclear genome. A biochemical analysis, the separation of the proteins via gel electrophoresis and/ or isoelectric focusing, reveals which protein is altered.
A third way to answer the question is the analysis of the protein biosynthesis in isolated chloroplasts or in in vitro systems assembled from chloroplast components. The finding that the oligomerous proteins of the photosynthesis membrane consist of subunits that are partially encoded by chloroplasts, partially by the nucleus is an interesting aspect. These complexes are good specimen for studying the interactions between both genomes.
The question, how such a co-operation has evolved remains unanswered. The endosymbiontic theory and several associated results of molecular biological studies ('jumping' or mobile genes) suggest that the whole required genetic information was originally located within the chloroplast DNA but that part of it migrated into the nucleus and has since been replicated and expressed just like any other nuclear gene. The following result may illustrate this: in the pea (Pisum sativum) are the gamma- and the delta- subunits of the ATPase synthesized within the nucleus, while the alpha-, beta- and the epsilon- subunits are produced in the chloroplasts (P. - Y. BOTHYETTE and A. T. JAGENDORF, 1978). In contrast synthesize the chloroplasts of spinach not only these three last mentioned subunits but also the gamma-subunit (N. NELSON, I. H. NELSON and G. G. SCHATZ, 1980).
How do the transport mechanisms of the chloroplast envelope work? A protein synthesized in the cytosol has to pass at least two membranes (the inner and the outer membrane). A few nuclear encoded proteins that act within the thylacoids, like plastocyanin, do even have to pass three membranes (S. SMEEKENS et al., 1985). At the outer chloroplast membrane are - at least in chloroplasts - no ribosomes found. Accordingly can no synthesis through the membrane exist. Transport has to occur after synthesis has been completed. B. DOBBERSTEIN, G. BLOBEL and N. - H. CHUA (Rockefeller University, 1977) found out that the proteins that are synthesized in the cytosol but designed for the transport into the chloroplasts are by far larger than their functional counterparts within the chloroplasts.
The additional part is termed transit sequence. It seems to recognize specific receptors at the outside of the chloroplast and specific carriers have to exist that bind to the proteins and transport them through the membrane. In addition is an endoprotease required that transfers the transport form of the protein into its active state. Such an enzyme was isolated from chloroplasts of Chlamydomonas reinhardii.
Chloroplasts that have been treated with a protease do not import proteins any more, i.e. the required receptors for the transfer sequences and the carriers have ceased to be in force and without them no protein can be transported.
The outlined mechanism does not describe, how the inner chloroplast membrane is crossed. This transport proved to be simpler than at first thought. R. DOUCE and his collaborators found out (1973) that the distances between the two membranes of the chloroplast envelope are not constant, but that both membranes oscillate. They touch each other at certain spots and at regular intervals where they fuse reversibly for a short period of time. A protein would only have to cross one membrane at such sites.
Many of the still open questions have been approached with the help of genetic engineering. Among other things was it found out that a so-called promiscuous DNA exists. The term marks DNA segments that occur in chloroplasts, mitochondria and the nucleus. Furthermore was it shown that the DNA of maize mitochondria contains the gene for ribulose - 1,5 - bisphosphate carboxylase (D. M. LONSDALE et al., 1983) and that ribosomal and messenger RNA from chloroplasts, mitochondria and Eschericia coli resemble each other (H. J. BOHNERT et al., 1980). Such result point at an exchange of genetic information between the single compartments of the cell.
Model of the interactions between plastid and nuclear encoded transcription and translation products TS: transit sequence: a N-terminal section of the polypeptide chain, essential for the penetration of the polypeptide across the membrane, subsequently being cleaved off proteolytically (according to W. BOTTOMLEY und H. J. BOHNERT, 1982).

The results above are what was known before the complete chloroplast DNA of two plant species was sequenced. In summer 1986 were the nucleotide sequences of the chloroplast DNA of Marchantia polymorpha (a liverwort) and Nicotiana tabacum published. Two Japanese research groups (K. OHYAMA and 12 collaborators, Kyoto University, and K. SHINOZAKI and 22 collaborators, Nagoya University) accomplished this work. The Marchantia - ctDNA contains 121,024, the tobacco - ctDNA contains 155,844 base pairs. Although the two genomes differ considerably in size are their sets of genes nearly alike, even the arrangement in almost unchanged. This result alone points conclusively at a common ancestor of the two chloroplast genomes. The Marchantia chloroplast genome contains presumably 128 genes, 4 of which are rRNA genes, 32 are tRNA genes and 55 are open reading frames (ORF). Open reading frames are nucleotide sequences that start with a start codon, end with a stop codon and are not interrupted by any further signal of this kind. Such sequences seem to be structural genes encoding proteins. Since an identified nucleotide sequence can without difficulties be translated into the respective amino acid sequence, were numerous protein genes identified. For example was a nucleotide sequence found that contains the information for a NADH dehydrogenase subunit so far only found in human mitochondria. This again indicates a promiscuous DNA.
The structure of the chloroplast DNA resembles that of a procaryotic DNA. Subsequent genes, for example, are transcribed en bloc (operon concept). In addition exist overlapping genes, i.e. nucleotide sequences that are part of two genes. For example is the sequence of gene 1
A - B - C - D - E - F
and of gene 2
E - F - G - H - I...
About 20 genes contain introns of differing lengths, a feature otherwise only found in eucaryotic DNA.
The chloroplast DNA of tobacco contains the gene for a certain ribosomal protein (S 16, i.e. protein number 16 of the small ribosomal subunit); this gene is missing in the ctDNA of liverwort. Instead contains the ctDNA of tobacco a gene for another ribosomal protein [L 21; i.e. protein number 21 of the large subunit], that is missing in the ctDNA of liverwort.
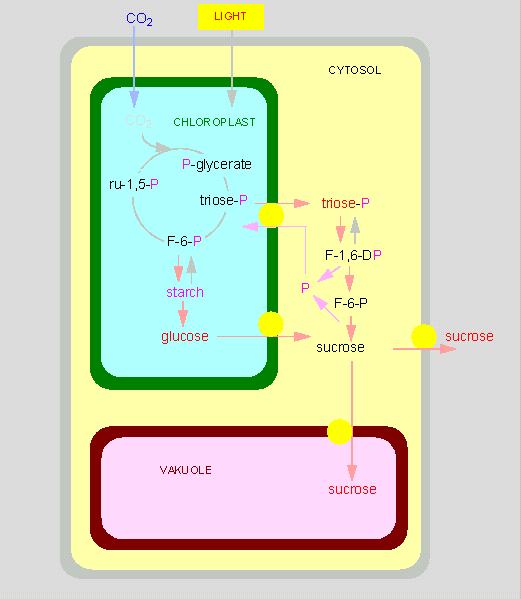
The chloroplasts are the site of photosynthesis and it is important to understand, which metabolites are taken up by the chloroplasts and which are secreted into the surrounding cytosol. Taken up are mainly water, carbon dioxide and phosphate. The reactions of the CALVIN cycle take place in the chloroplast stroma. The final product glucose is transported into the cytosol where it is used for the synthesis of saccharose that is either stored within the vacuole or exported out of the cell. Besides saccharose are primarily the triosephosphates dihydroxy acetonephosphate and glycerine aldehydephosphate put at the cell's disposal. The triosephosphate transport and the import of the phosphate are energy-consuming and occur with the help of a phosphate transporter.
© Peter v. Sengbusch - Impressum